FAQs
Scope
This article includes frequently asked questions, organized by topic.
Disk Use
Question: Can only one disk be associated with one workflow?
Yes. That disk is local to that workflow, as it is an attached drive to that workflow.
Question: What is the relationship between disk and file sizes?
The larger the disk, incrementally the more throughput you’ll get. So, if we compare a 100 GB disk with a 1 TB disk, the 1 TB disk is not just 10x bigger, it also is faster.
It’s helpful to think about how your data set is organized. When it comes to file sizes and interactions, there are differences in disks that need to read 100 MBs from 1 file vs 10 MBs from 10 files. Reading from multiple smaller files can be slower than reading from one large file.
Question: What is the ideal disk space?
As a best practice, your data set should represent about 75% of the disk space you have available. We recommend a default of 100 GB of disk space. Disk space is relatively inexpensive (pennies per gigabyte per month). The nature of the analysis can impact the choice of disk size. For example, some NONMEM analysis may not inherently use much disk space per run, but if you run a bootstrap scenario with 1000 models, each taking up 10 MB of space, you may find that your simulation has now taken up 10 GB.
Question: How can I determine my disk space utilization?
One way is to look at the disk utilization indicator on your workflow dashboard.
Another way to check resource utilization is via Grafana. Simply change your workflow URL so that it ends in /grafana.
Question: After I initially create a disk, can I increase the size?
Yes, this can be done when creating a new workflow. For more information, please see Working with Disks.
Question: Will all data get carried over after the disk size is changed?
Yes. Everything that was on that disk will be available.
Question: Can I delete a disk that is no longer being used?
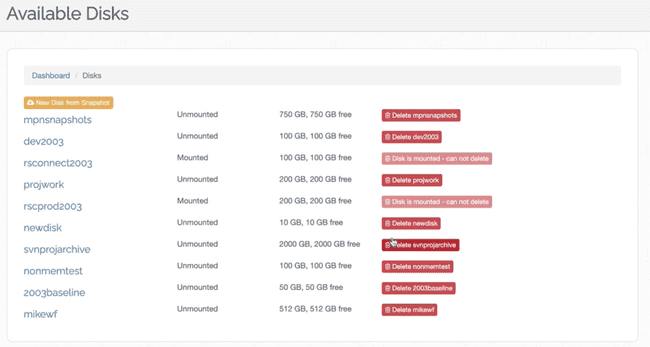
Yes. Your organization Administrator can do this, or you can do it yourself. On the Metworx dashboard, click on your user profile, then click My Disks
- Then, you will see all of your available disks. Any disk that is not currently mounted, you can delete.
Saving Work
Question: What happens if two worker nodes are writing to the same file? What happens if there is a conflict?
Two compute nodes can write to the same file at the same time without issue.
However, further consideration is needed when running two analyses (or parallelized analyses) on your head node. If you have two analyses running at the same time, and you attempt to save each to the same file, one will overwrite the other. In this scenario, as a best practice, you should use some additional file prefix or suffix to delineate between them, then subsequently you can aggregate those to a single result/file.
For example, if you have four processes running, rather than saving everything out to something like “results.csv”, you can alternatively write out to “resultsp1”, “resultsp2”, etc. After your analyses jobs have completed, you could aggregate the files in a final results file.
Question: What if a user has 10,000 compute nodes that may be writing independent files but are all trying to write to /data simultaneously?
Users can stream out a pretty significant amount of data.
There have been times where MetrumRG’s internal science group has spun up a 9600 core workflow that was running 9600 models at a time, and each of those models was writing out 10-20 files at a time with no problem on a two-core master node.
Question: If I have a file in my /tmp file on a compute node, and I stop my workflow, will that file be erased?
Yes, that file will be erased. As a best practice, it’s better to store your work in /data on the head node. Anything put in /data on your head node will become instantly available in your compute nodes as well as via the /data file mount.
/tmp is the local disk to that compute node. The compute node also has a network file mount to /data from the head node. So, essentially the head node acts as a file share for all of the compute nodes via /data.
Question: I have a 1 GB file saved in /data on my head node. If I then have 2 compute nodes, will the file be shared across the 2 compute nodes evenly (so 500 MB on both?)
No, what will happen is that the file will be fully available on both compute nodes at the same time because /data is a network file share.
If you have a script that needs to read a file from /data on a compute node, that script will be able to access the entirety of that data file.
If you need to do distributed processing across multiple compute nodes in a manner that the first 500 MB is run on one compute node, and the second 500 MB on a second compute node, then that is an exercise of writing your scripts such that they account for this distribution, for example:
- On the first compute node, the script reads in the entire file from /data then discards the second half of it.
- On the second compute node, the script reads in the entire file from /data then discards the first half of it.
RStudio Sessions
Question: Does the RStudio session require a workflow to run? If not, then why is the RStudio session state saved? For example, after wrapping up an RStudio session and shutting down the associated workflow for the day, if the next day I start up a new workflow, I notice that when I load RStudio, whatever state the session was in on the previous day persists to the next day.
Yes, the RStudio session does require a workflow to run.
The RStudio session is stored on the user's disk. Because your disk is in your home directory, when a workflow shuts down, it automatically backs up everything under /data. A snapshot is taken of your disk and backed up. What this means is that the next time you attach it to a new workflow, the disk that is reattached is a literal snapshot in time of exactly where you left off.
Question: Can I change the RStudio session default settings?
As a best practice, we recommend using new RStudio sessions each time a new workflow is launched so you do not accidentally introduce some sort of state to your analysis activities.
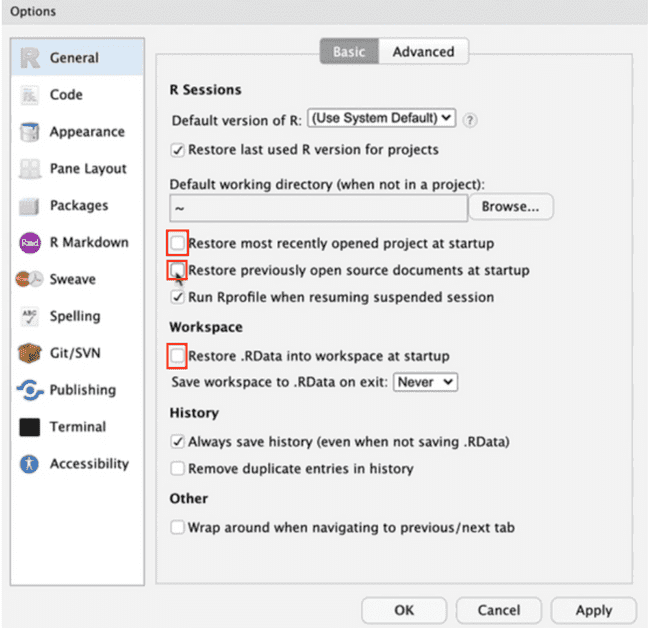
If you do not like the default behavior:
- Go to Global Options
- In the "Basic" section of the General R options, change the configuration to your preference:
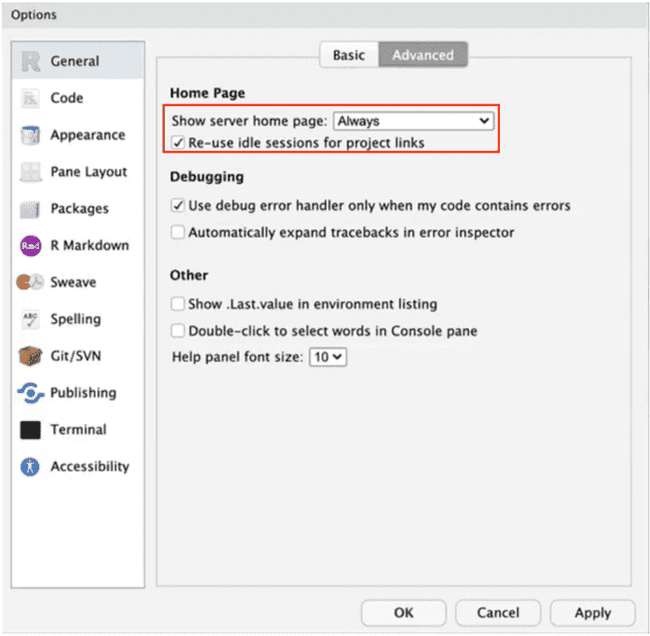
- In the "Advanced" section of the General R options, make sure "Show server home page" is set to "Always".
When doing this, you will be taken to the RStudio start page where you will be able to see your previous sessions. Your sessions will be cleared out due to the changes you made in the "Basic" section above.
Remember, if you choose to set up RStudio so sessions are not saved, the files in your disk will persist across workflows. However, it’s important to understand they will not persist in the specific state associated with your terminal sessions.
Desktop Console
Question: Are there other ways to start up a compute node?
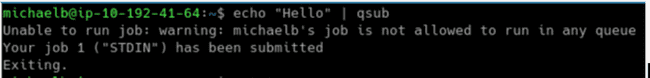
To get a compute node to start up, you can submit a fake job:
$ echo "Hello" | qsubQuestion: How can I check the number of queued jobs and node stat usage from the desktop console?
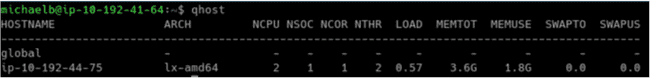
Run 'qhost' :
$ qhost